
Introduction
We tested SARS-CoV-2 primers from three different groups, 3 RT-qPCR kits and 3 RNA extraction kits to identify whether there was any difference in performance. The cost of these reagents were also compared, so that we could make cost-performance analysis.
Product choice
We chose to test these products because, at the time of study planning, when we contacted the companies, these were all available with a short lead time. Due to shortages of reagents and consumables relating to COVID-19 diagnostics, many items were unavailable or had long delivery times. The availability of these items may be different depending on your geographic location.
Due to the unavailability or delay in delivery time for commercial SARS-CoV-2 PCR assays (in which PCR reagents and primers are included in one kit), we did not include these in our testing.
The primers we tested were from:
- The Charite protocol
- The CDC protocol
- The Health Services Laboratories (HSL) protocol
RT-qPCR kits tested:
- Promega GoTaq® Probe 1- Step RT-qPCR System
- Takara One Step PrimeScript III RT-PCR Kit
- Thermo Fisher TaqMan™ Fast Virus 1-Step Master Mix
Viral RNA extraction kits tested:
- Quick-RNA Viral Kit w/ Zymo-Spin™ IC Columns (Capped)
- Qiagen QIAmp viral RNA miniprep kit
- Sigma GenElute™ Total RNA Purification Kit
SARS-CoV-2 RNA standard used
The standard used was donated to us by the LCG group. The standard was a synthetically produced 2938 nucleotide strand of RNA containing includes the N, RdRp and E genes of the SARS-CoV-2 virus. Digital PCR was used to quantify the standard and it was provided to us at 1x10^5 copies/µl.
Primers
We ordered the primers listed in Table 1 from Eurofins.
Methods
The SARS-CoV-2 standard was serially diluted from 1x10^5 to 1x10^0 and 5µl used in each reaction according to the PCR kit manufacturers instructions. All primers were testes with all 3 RT-qPCR kits.
Table 1. Primer names, their source and sequences.
|
Source |
Primer name (probes) |
Sequence (5’ to 3’) |
| Charité protocol. Details in Corman et. al (2020) |
E Sarbeco |
|
|
E_Sarbeco_F1 |
ACA GGT ACG TTA ATA GTT AAT AGC GT |
|
|
E_Sarbeco_R2 |
ATA TTG CAG CAG TAC GCA CAC A |
|
|
E_Sarbeco_P1 (5'-FAM/BBQ-3') |
ACA CTA GCC ATC CTT ACT GCG CTT CG |
|
|
N Sarbeco |
||
|
N gene N_Sarbeco_F1 |
CAC ATT GGC ACC CGC AAT C |
|
|
N_Sarbeco_R1 |
GAG GAA CGA GAA GAG GCT TG |
|
|
N_Sarbeco_P1 (5'-FAM/BBQ-3') |
ACT TCC TCA AGG AAC AAC ATT GCC A |
|
|
RdRp |
||
|
RdRP_SARSr-F2 |
GTG ARA TGG TCA TGT GTG GCG G |
|
|
RdRP_SARSr-R1 |
CAR ATG TTA AAS ACA CTA TTA GCA TA |
|
|
RdRP_SARSr-P1 (5'-FAM/BBQ-3') (Pan Sarbeco-Probe) |
CCA GGT GGW ACR TCA TCM GGT GAT GC |
|
|
CDC2
|
N1 gene |
|
|
2019-nCoV_N1 F |
GAC CCC AAA ATC AGC GAA AT |
|
|
2019-nCoV_N1-R |
TCT GGT TAC TGC CAG TTG AAT CTG |
|
|
2019-nCoV_N1-P (FAM/BHQ-1) |
ACC CCG CAT TAC GTT TGG TGG ACC |
|
|
N2 gene |
||
|
2019-nCoV_N2-F |
TTA CAA ACA TTG GCC GCA AA |
|
|
2019-nCoV_N2-R |
GCG CGA CAT TCC GAA GAA |
|
|
2019-nCoV_N2-P (FAM/BHQ-1) |
ACA ATT TGC CCC CAG CGC TTC AG |
|
|
HSL
|
N-gene |
|
|
NgeneTaq1 |
TCT GGT AAA GGC CAA CAA CAA |
|
|
NgeneTaq2 |
TGT ATG CTT TAG TGG CAG TAC G |
|
|
NgeneProbe (FAM/BHQ-1) |
CTG TCA CTA AGA AAT CTG CTG CTG AGG C |
|
Primer results
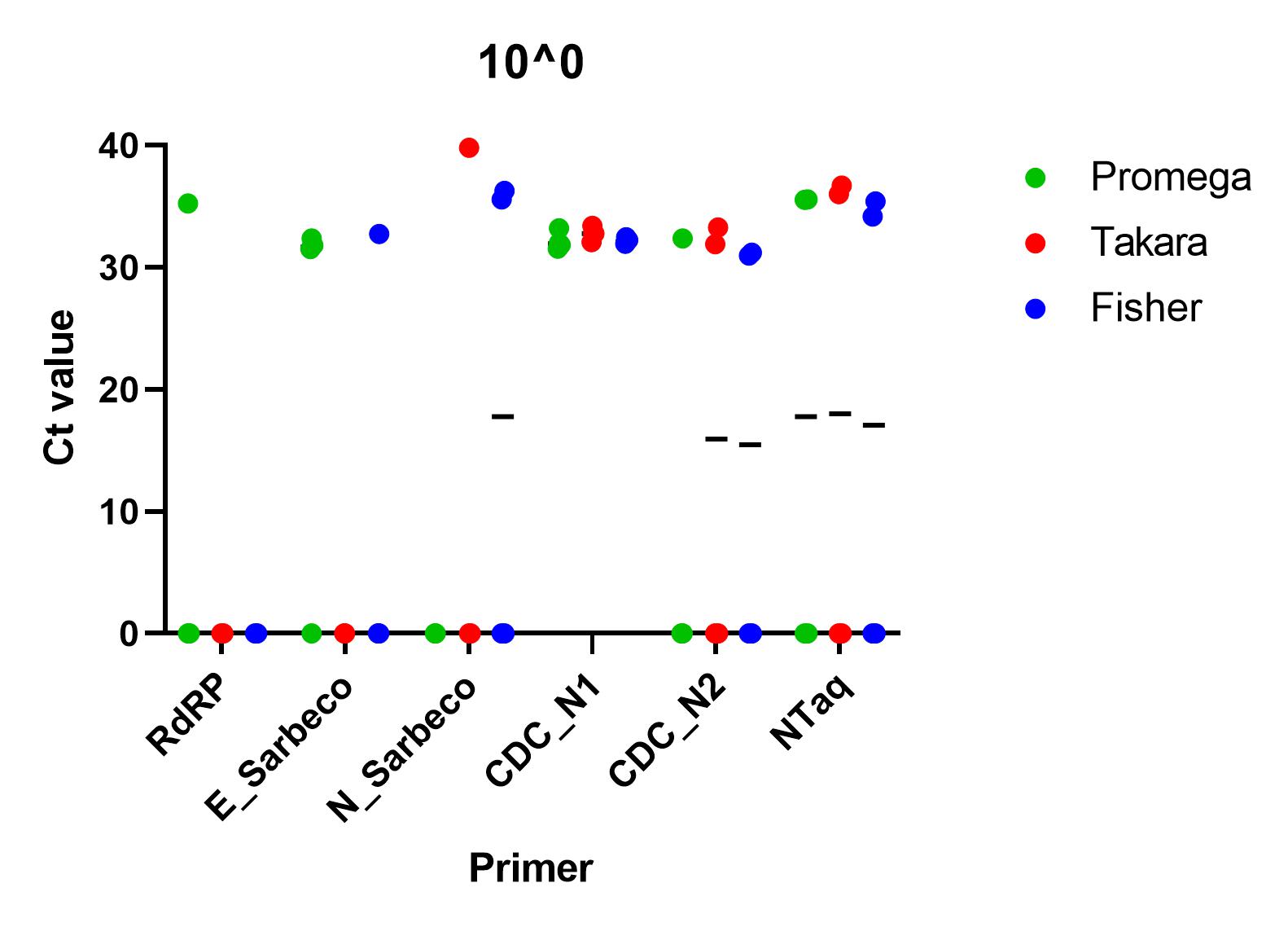
When the primers were tested using a SARS-CoV-2 standard serially diluted from 1x10^4 to 1x10^0, we found that all of the primer sets successfully detected nucleic acid down to 1x10^1. At lower concentrations than this, all primers, apart from CDC’s N1 primer set, failed to detect in all repeats. At all other concentrations, there was a significant difference between the ct values (p=0.0001), using one way ANOVA and the Charité E_Sarbeco, CDC N1 and CDC N2 primer sets performed better.
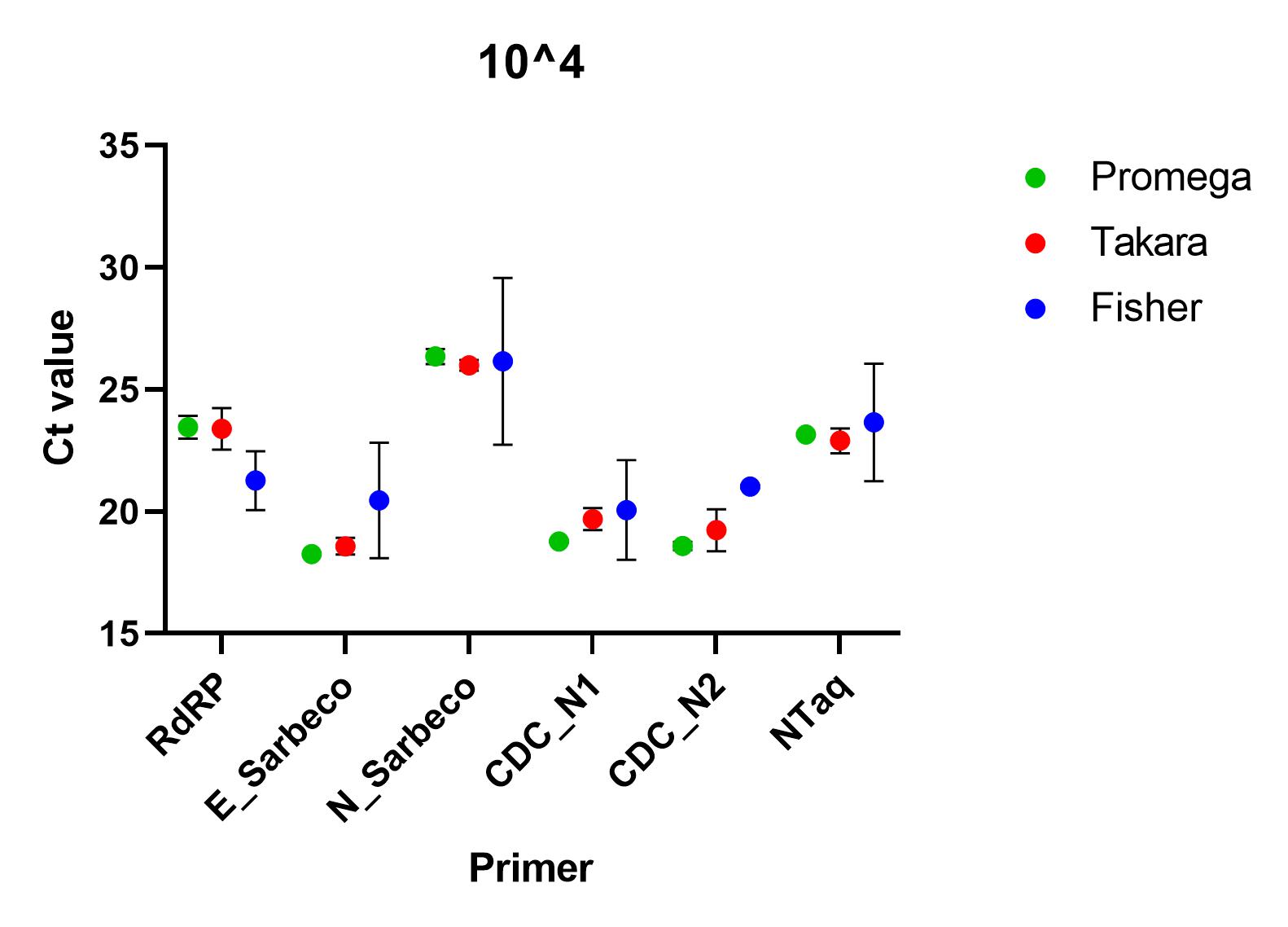
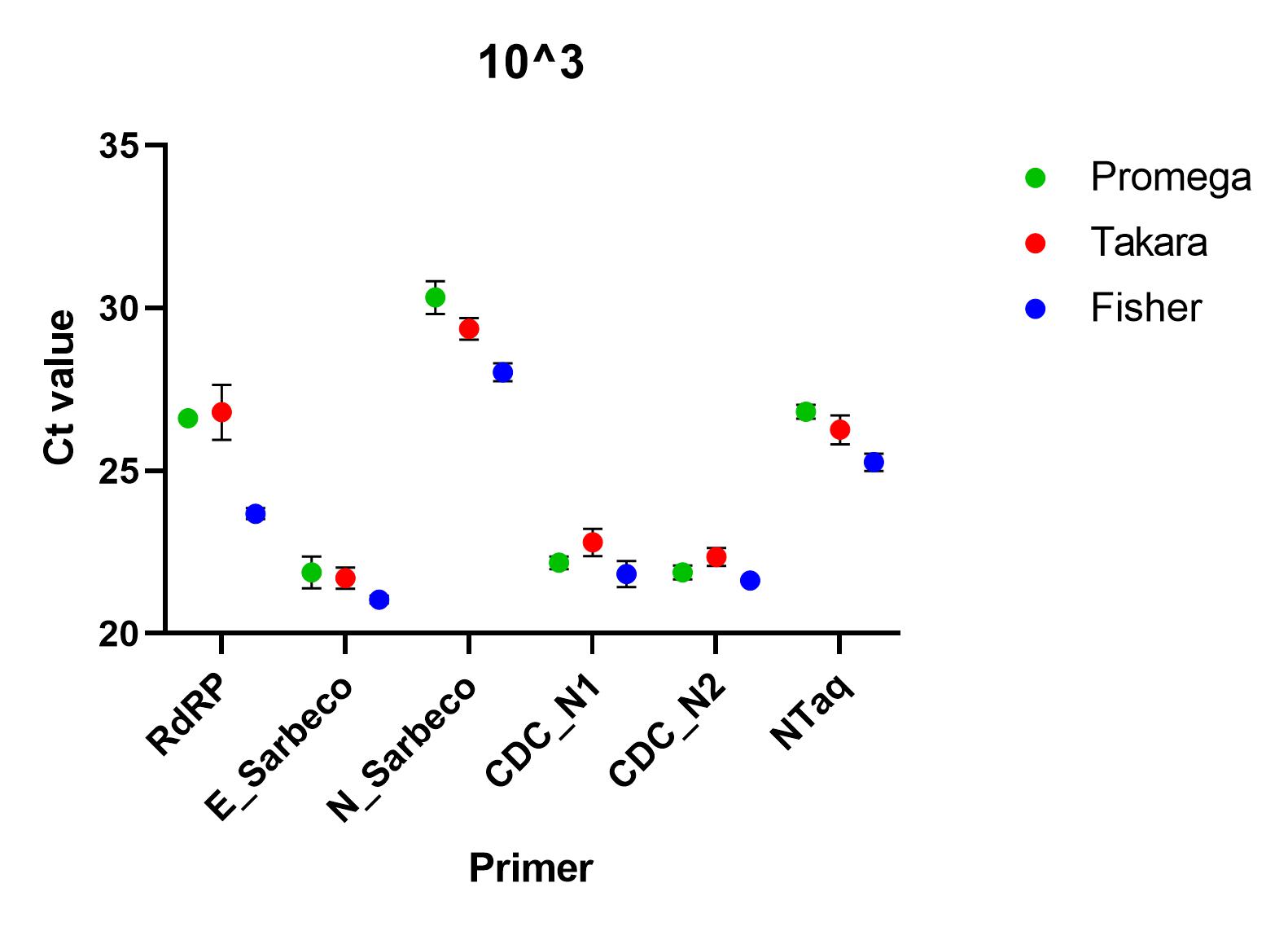
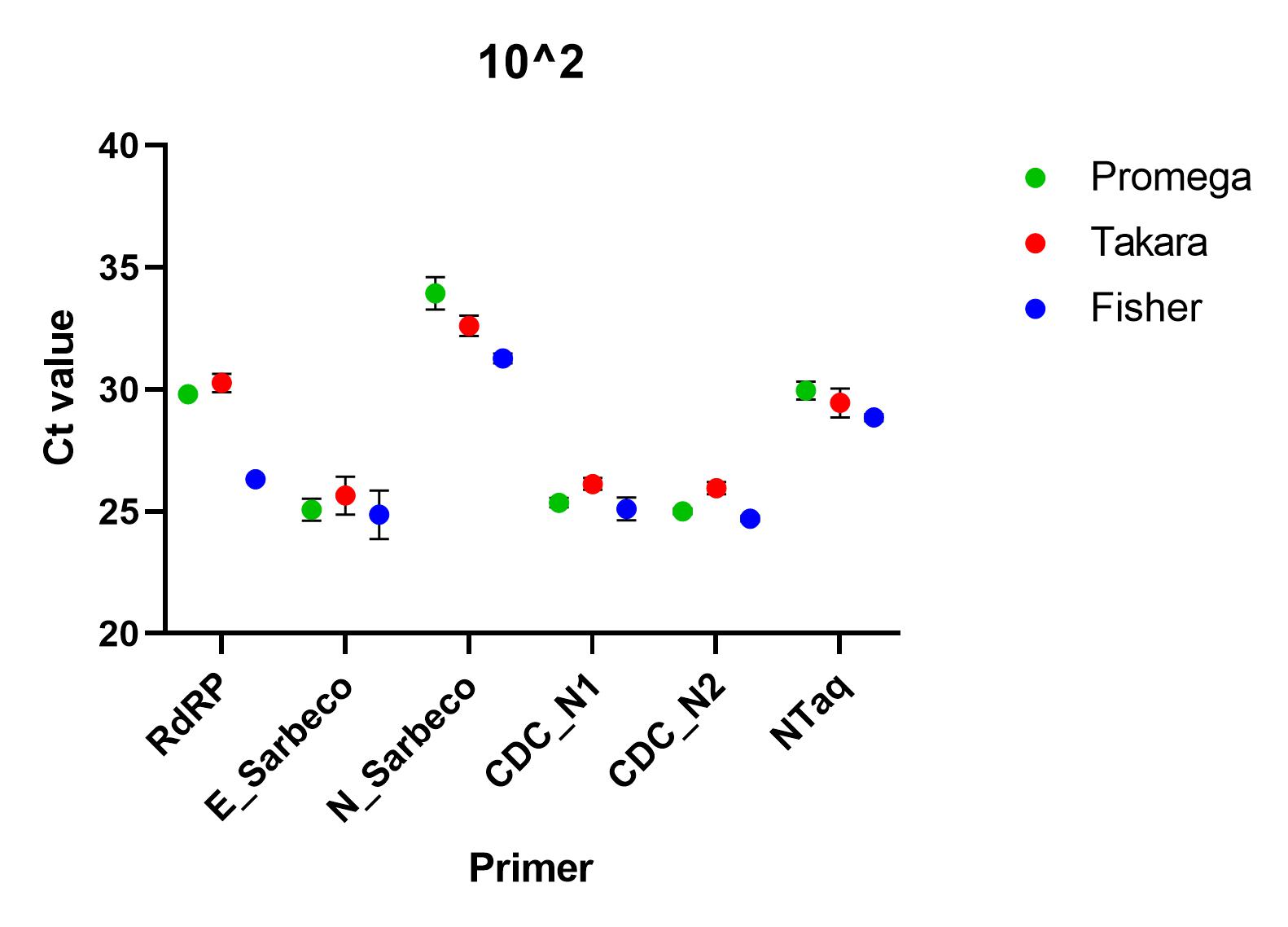
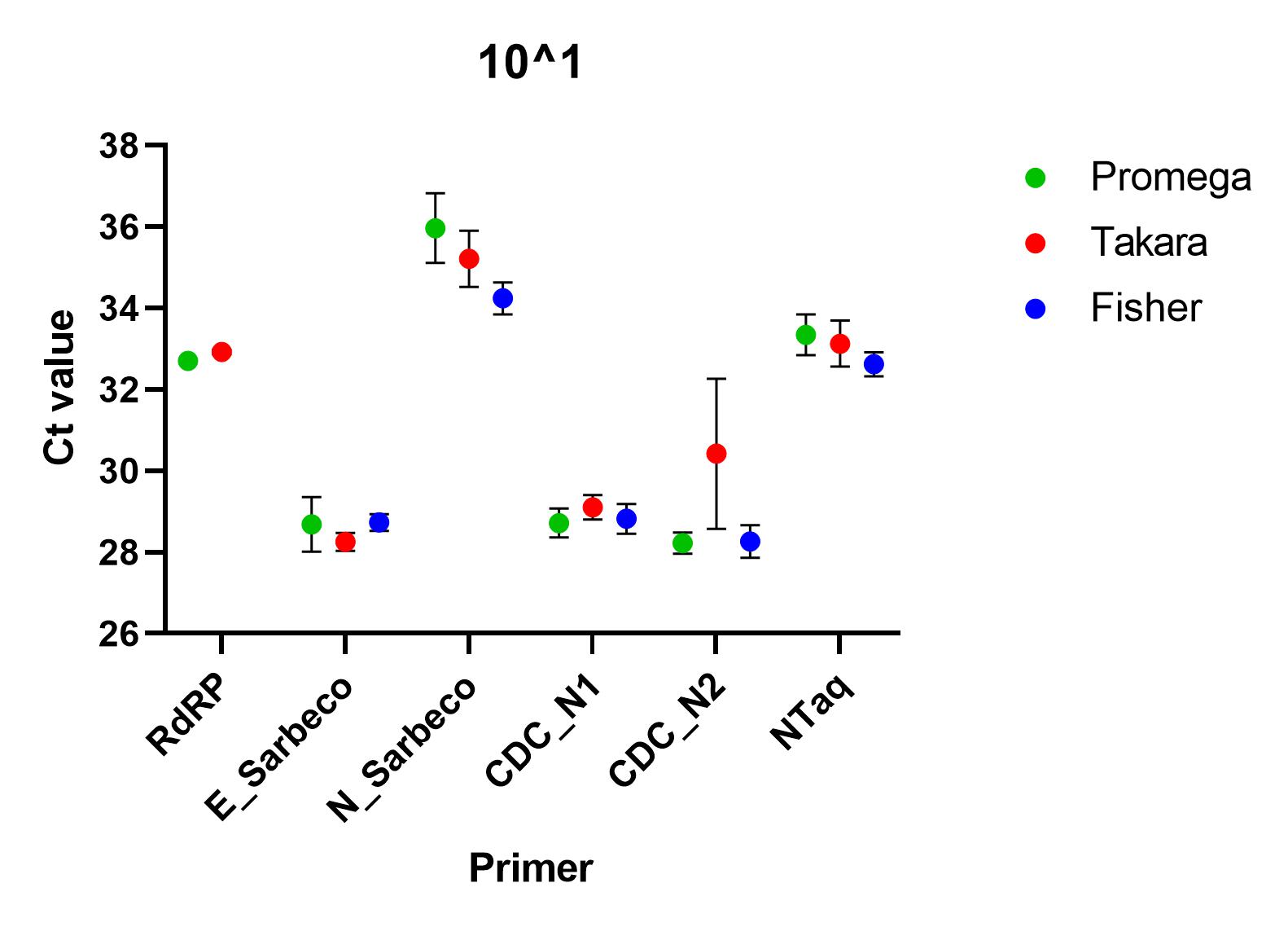
Recommendations
All of the primer sets we tested could reliably detect SARS-CoV-2 nucleic acid to 1x10^1 copies/µl. Charité’s E sarbeco, and CDC’s N1 and N2 performed better than Charité’s RdRp, N Sarbeco and HSL’s NTaq primer sets. For samples of less than 1x10^1 copies/µl, we recommend concentrating samples prior to extraction. Note that for Charité’s RdRP primer set, low sensitivity may be due to mismatch in R primer.
RT-qPCR systems
Method
We chose to test one-step kits rather than two-step kits due to ease and speed of PCR preparation (important in high throughput scenarios such as diagnostics). The kits tested are designed to work with the above mentioned primers, which utilise fluorescent probes, rather than dyes, such as SYBR Green. We tested each RT-qPCR kit according to the manufacturers preparation and cycle condition protocols. See table 2 for kit product codes, list prices and links to the manufacturers website. Prices are correct as of 31st March 2021. All costs are in GBP.
NB it is important to check which thermocycler will work with these PCR systems, this information can usually be found in the kit manuals.
Table 2. RT-qPCR kit information
|
RT-PCR system |
Manufacturer |
Product code |
Number of reactions per kit |
Website list price (GBP) (31 Mar 2021) |
Cost per reaction (GBP) |
Website address
|
Kit manual |
|
GoTaq® Probe 1- Step RT-qPCR System |
Promega |
A6120 |
200 |
£234.00 |
£1.17 |
||
|
One Step PrimeScript III RT-PCR Kit |
Takara |
RR600 |
200 |
£293.00 |
£1.47 |
||
|
TaqMan™ Fast Virus 1-Step Master Mix |
Thermo Fisher |
4444432 |
200 |
£264.00 |
£1.34 |
Results
The results shown here include our results using the three best performing primer sets, Charite's E sarbeco, and CDC’s N1 and N2.
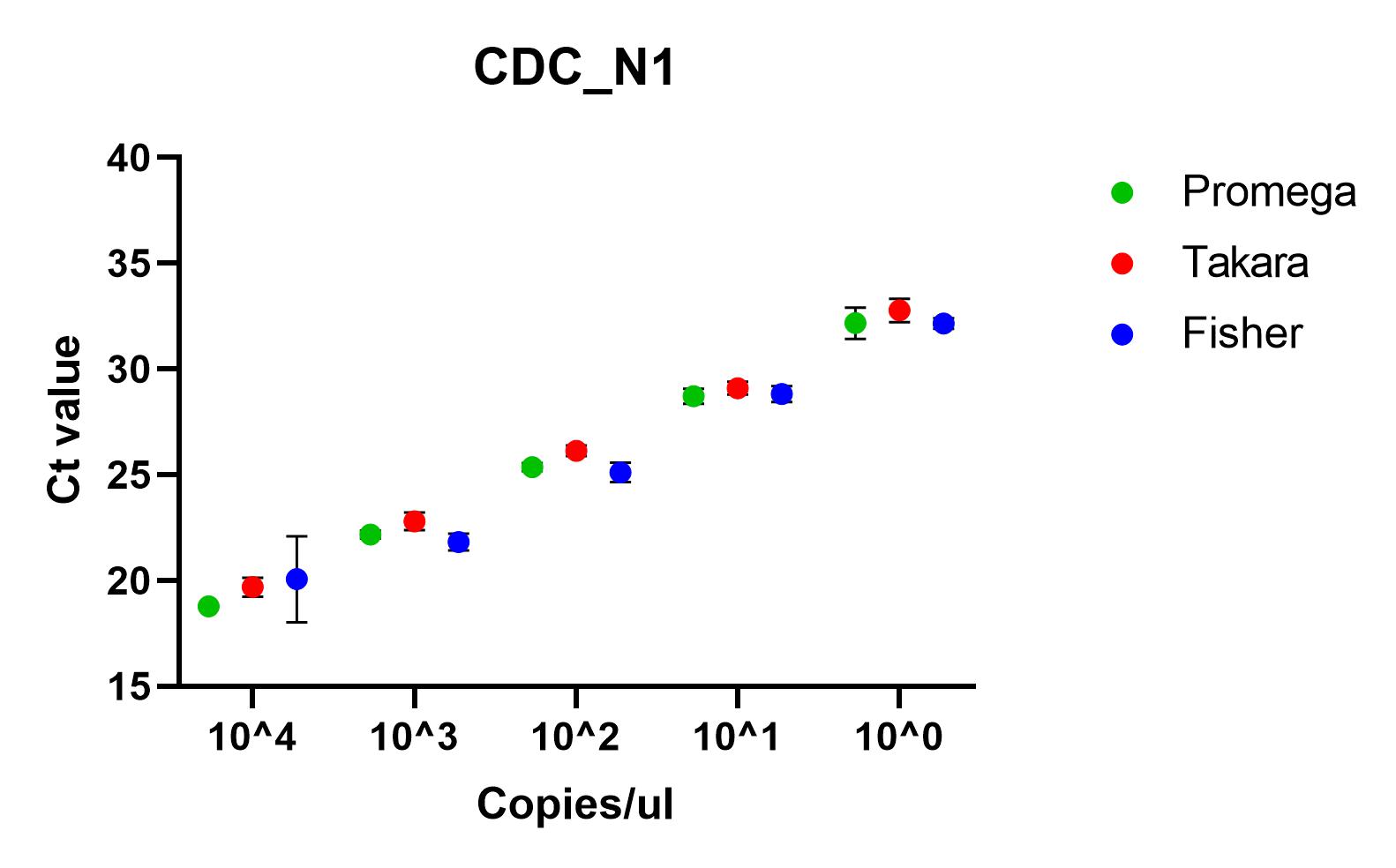
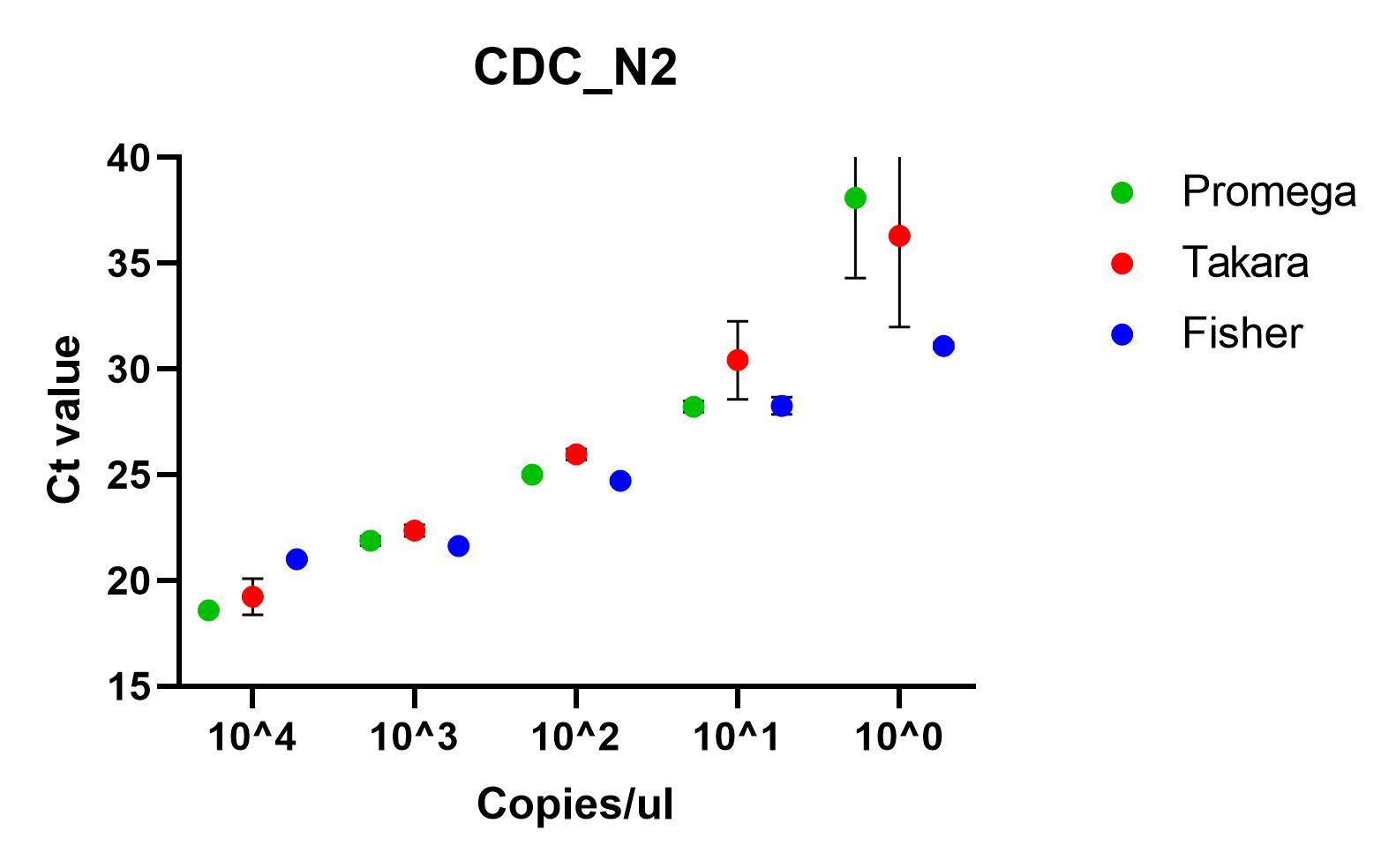
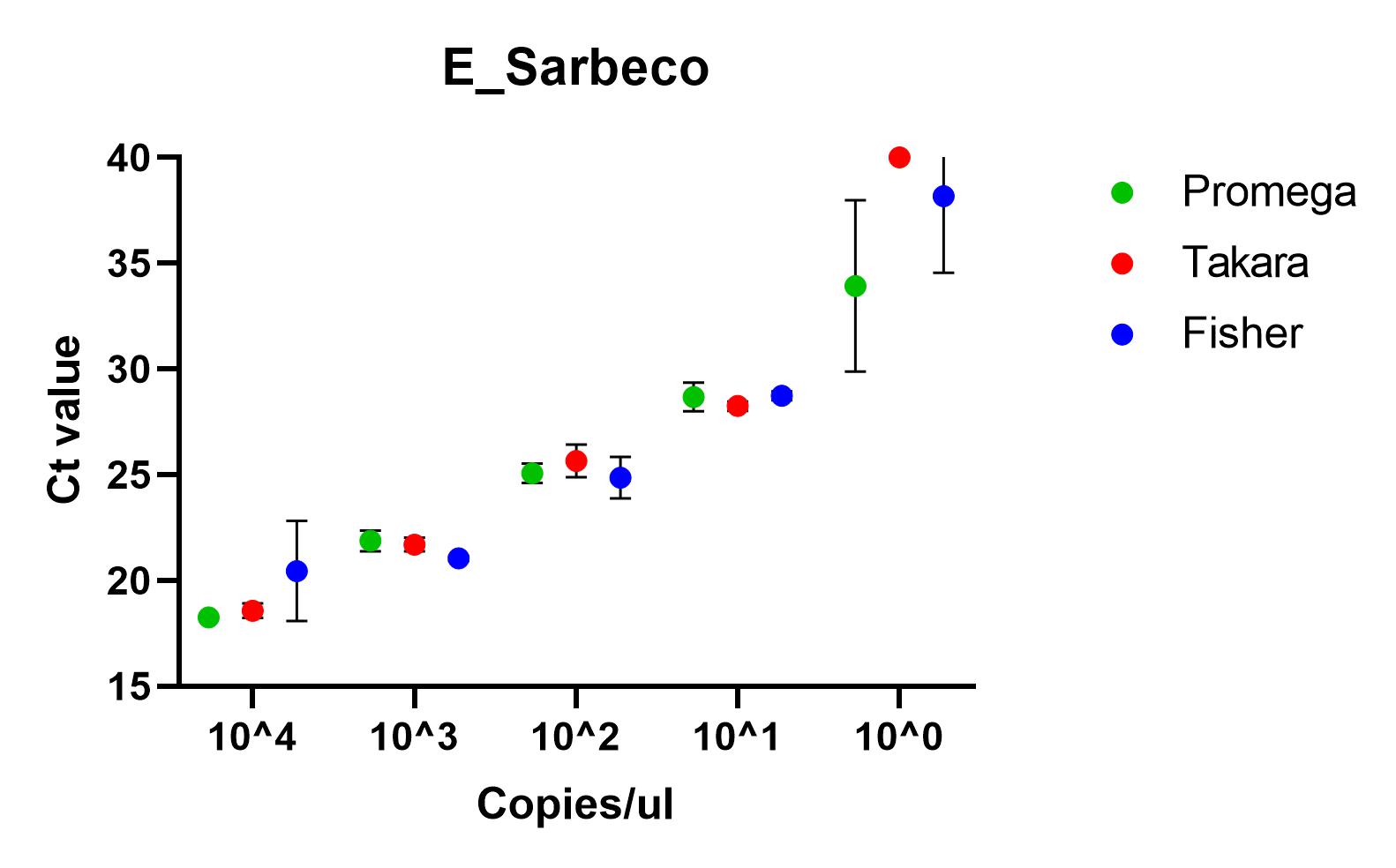
Recommendations
All of the primer sets we tested could reliably detect SARS-CoV-2 nucleic acid to 1x10^1 copies/µl. Charité’s E sarbeco, and CDC’s N1 and N2 performed better than Charité’s RdRp, N Sarbeco and HSL’s NTaq primer sets. For samples of less than 1x10^1 copies/µl, we recommend concentrating samples prior to extraction. Note that for Charité’s RdRP primer set, low sensitivity may be due to mismatch in R primer.
Viral RNA extraction kits
We tested each viral RNA extraction kit according to the manufacturers protocols. See table 3 for extraction kit product codes, list prices and links to the manufacturers website. Prices are correct as of 31st March 2021. All costs are in GBP.
Table 3. Extraction kits used and their manufacturer information
|
Extraction kit name |
Manufacturer |
Product code |
Extractions per kit |
List price per kit (GBP) (31 March 2021) |
Cost per extraction (GBP) |
Product website |
Kit manual |
|
Quick-RNA Viral Kit w/ Zymo-Spin™ IC Columns (Capped) |
Zymo |
R1034 |
50 |
£172.00 |
£3.44 |
||
|
QIAmp viral RNA miniprep kit |
Qiagen |
52904 |
50 |
£214.00 |
£4.28 |
||
|
GenElute™ Total RNA Purification Kit |
Sigma |
RNB100-50RXN |
50 |
£410.00 |
£8.20 |
Method
Note that as we were testing SARS-CoV-2 serially diluted from cultures, and extracting from high-concentration samples, all work was carried out under BSL3 conditions. SARS-CoV-2 is classified as Hazard Group (HG) 3 organism when cultured. It is classed as a HG2 organism and therefore can be processed in a BSL2 laboratory using a microbiological safety cabinet and the appropriate PPE if working on samples. Please refer to our COVID-19 diagnostics health and safety page for more information and links to resources.
Swab method
We serially diluted a 8x10^5 PFU/µl culture of SARS-CoV-2 to 8x10^1. In an Eppendorf, 45µl of SARS-CoV-2 culture was applied to a swab, which was then immediately transferred to a screw cap tube containing 600µl DNA/RNA shield and then frozen at -20°C until it was extracted.
We extracted from swabs containing 10^3, 10^2 and 10^1 SARS-CoV-2 culture dilutions. 100µl of the media surrounding the swab was then extracted with each kit following the manufacturers instructions. Each dilution for each kit was repeated 3 times. 5µl of eluted RNA from each kit was then tested using the CDC N1 primer set and TaqMan™ Fast Virus 1-Step Master Mix (following manufacturers instructions).
Use of DNA/RNA shield
When we requested information from the suppliers of the extraction kits regarding validation of viral inactivation, only Zymo told us that their inactivation step, utilising the DNA/RNA Shield product, had been validated. Therefore the other kit manufacturers could not guarantee that the virus had been inactivated using their kits. Because of this, and UCL risk assessments, we placed all of our swabs directly into 600µl DNA/RNA shield before proceeding with each of the extraction kits.
Results
One way ANOVA showed no significant difference between the tests at any of the concentrations, suggesting that, when following manufacturers instructions, each of the viral RNA extraction kits tests should work equally well.
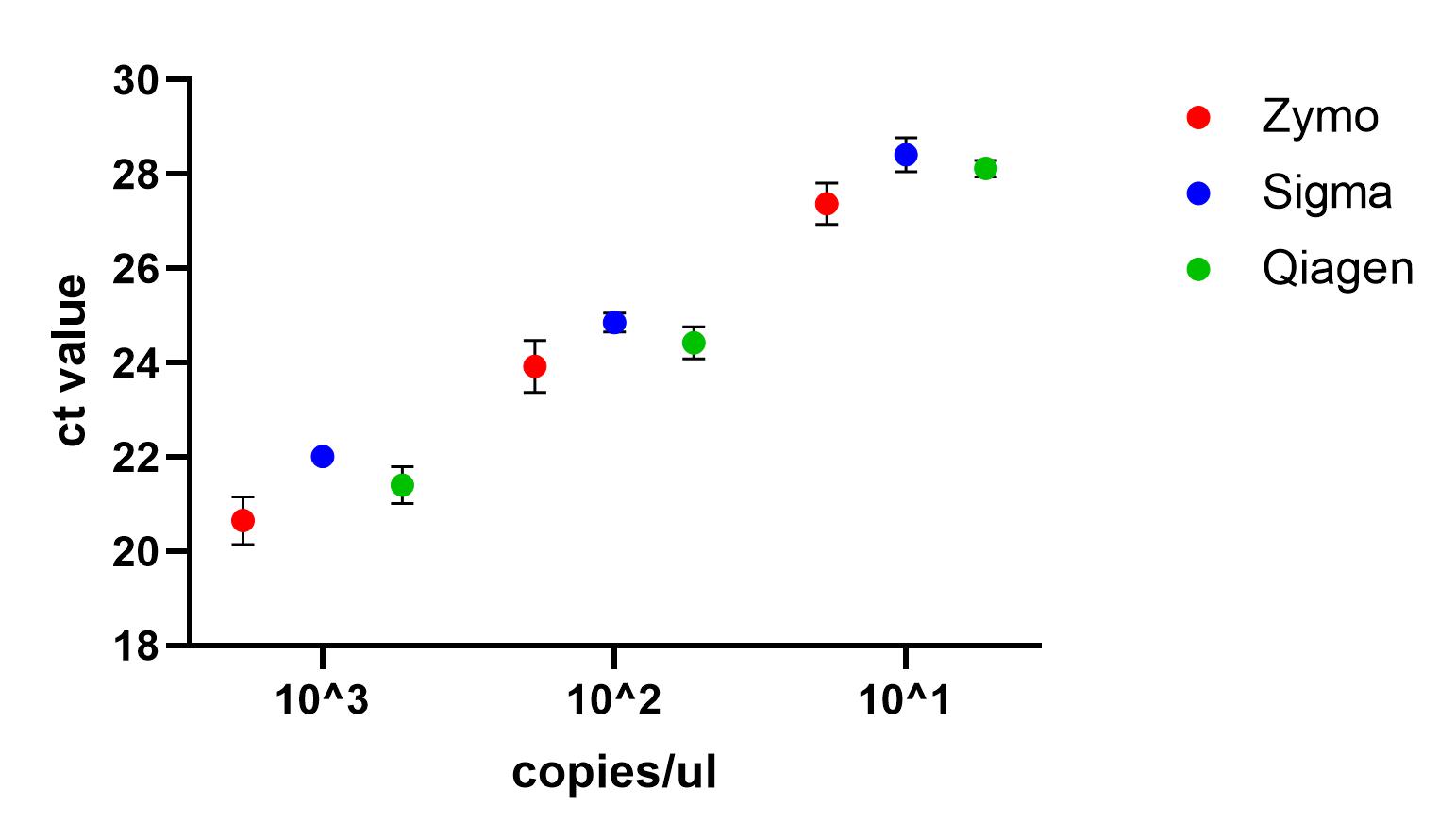
Recommendations
All three kits can detect SARS-CoV-2 nucleic acid down to 1x10^1 copies/µl, although the Zymo kit appears to perform slightly better, based on a visual inspection of the data.
Cost review
We have created a cost matrix to identify which combination of RT-qPCR kits and viral RNA extraction kits are cheapest (the below costs do not include plasticware and other consumables not provided with the kits). Tables 4 and 5 show that the combination of the Promega RT-qPCR kit and Zymo extraction kit is the cheapest, coming out at £4.61 per sample. If utilising DNA/RNA Shield in addition to the Qiagen or Sigma extraction kits, the cost will increase, but it has been validated to inactivate SARS-CoV-2 (table 5).
Table 4. Cost matrix (without addition of DNA/RNA shield to Qiagen and Sigma extraction kits)
|
Promega |
Takara |
Fisher |
|
|
Zymo |
£4.61 |
£4.91 |
£4.78 |
|
Qiagen |
£5.45 |
£5.75 |
£5.62 |
|
Sigma |
£9.37 |
£9.67 |
£9.54 |
Table 5. Cost matrix (with addition of DNA/RNA shield to Qiagen and Sigma extraction kits)
|
Promega |
Takara |
Fisher |
|
|
Zymo |
£4.61 |
£4.91 |
£4.78 |
|
Qiagen |
£7.01 |
£7.31 |
£7.18 |
|
Sigma |
£10.93 |
£11.23 |
£11.10 |
Overall recommendations
Our overall recommendations:
- In terms of the RT-qPCR step, it seemed that the choice of primer affected the results more than the kits that were tested. The Charité E sarbeco, and CDC’s N1 and N2 primer sets performed significantly better than Charité’s RdRp, N Sarbeco and HSL’s NTaq primer sets.
- There was no significant difference in the performance of the RT-qPCR kits tested.
- All three of the extraction kits detected SARS-CoV-2 to 1x10^1 copies/µl and there was no significant difference between them.
- In terms of cost analysis, the Promega RT-qPCR kit and Zymo extraction kit came out as the cheapest combination, at £4.61 per sample (not including plastic ware etc.).
- The most expensive combination was the Takara PCR kit and Sigma extraction kit, which at £11.23, was more than twice as expensive.
- Given that both the PCR kits and the extraction kits performed similarly, availability and choice of primers should be the most important decisions when setting this assay up.